
Discovery of a novel coral bacterium with a potent ability of releasing climate-cooling gase, dimethylsulfide.
DMSP (dimethylsulfoniopropionate) is an important global sulfur cycle molecule, primarily produced by marine phytoplankton and microorganisms. Coral reefs are habitats with high DMSP production, where not only planktonic microorganisms generate this molecule, but the corals themselves can also produce DMSP. The metabolism of DMSP is considered one of the most crucial mechanisms and means for corals to counteract heat stress. Among its breakdown products, DMS (dimethylsulfide) is a well-known climate-cooling gas with potent antioxidative abilities, aiding corals in clearing excessive free radicals induced by heat stress.
The primary metabolism of DMSP into DMS is largely performed by prokaryotic microorganisms. Hence, whether dominant symbiotic bacteria in corals possess the ability to decompose DMSP has been a long-standing concern. Endozoicomonas, an endosymbiotic bacterial genus commonly found in corals, was observed to potentially play a role in the breakdown of DMSP and DMS production in corals. However, questions persisted regarding whether E. acroporae was merely an isolated case and whether the decomposition of DMSP and DMS production within coral organisms represents a crucial ecological function of Endozoicomonas. Mechanisms related to molecular regulation remained unknown.
In this study, we successfully isolated a new strain of endosymbiotic bacteria, named Endozoicomonas ruthgatesiae 8E, and through genomic and biochemical analysis, confirmed that E. ruthgatesiae possesses key DddD enzymes and the related metabolic gene cluster similar to those in E. acroporae for the breakdown of DMSP and the production of DMS. Utilizing bioinformatics analysis, we also found several crucial dddD genes in the genomes of several published but yet-to-be-cultivated Endozoicomonas, indicating that the ability to decompose DMSP is rather common within the genus of endosymbiotic bacteria, rather than being only restricted to a few bacterial species.
Furthermore, through comparative biochemical and transcriptome analyses, we discovered that although E. ruthgatesiae and E. acroporae possess similar genomic characteristics in the dddD gene group, the ways in which the DddD enzyme induces the metabolism of DMSP differ. E. ruthgatesiae rapidly absorbs DMSP, simultaneously rapidly producing DMS without accumulating it within the organism. Conversely, E. acroporae quickly absorbs DMSP but accumulates it, subsequently releasing DMS slowly. This distinct biochemical reaction was also confirmed in the comparative transcriptome analysis. E. ruthgatesiae was upregulating the DMSP metabolic genes and concurrently regulating many energy-related metabolic genes. However, E. acroporae was upregulating DMSP metabolism while negatively regulating energy-related metabolism. This indicates that although both strains of bacteria accommodate a similar dddD gene cluster, the way they use DMSP exhibits different genetic regulations and biochemical reactions. This reveals that the DddD-induced DMSP metabolism mechanism presents variations. This is the first demonstration of the gene regulation and analysis of DddD-induced DMSP breakdown and also the first instance to present the diverse characteristics of DMSP metabolism among endosymbiotic bacteria and their ecological functional differences.
The species name "Endozoicomonas ruthgatesiae" is named in honor of Dr. Ruth Gates from the University of Hawaii, who has long dedicated herself to the conservation and restoration of coral reefs (https://en.wikipedia.org/wiki/Ruth_Gates).
The results were published on Science Advances. The link of the paper is: https://www.science.org/doi/10.1126/sciadv.adk1910
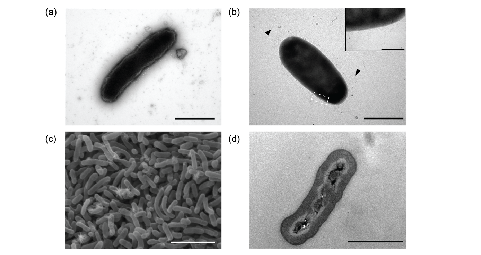
Fig 1. Electronic micrographs of Endozoicomonas ruthgatesiae. (a)(b) negative staining transmission electronic micrographs; (c) scanning electronic micrograph; (d) thin-section transmission electronic micrograph.
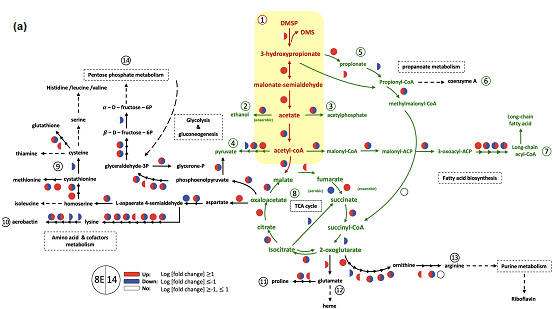
Fig 2. Comparative transcriptomic analysis of the both Endozoicomonas species. Yellow-highlighted area is the metabolic pathway of DMSP; the both bacteria upregulated the genes of the pathway (red). In the energy-related metabolisms, most of the genes are upregulated in E. ruthgatesiae but downregulated in E. acroporae (blue) indicating that there are variations of DMSP metabolism in the both bacteria.





